5/7/2019
Identifican patrones en tuits en español que pueden indicar signos de depresión
El estudio del grupo de Investigación en Informática Biomédica Integrada ha analizado los patrones lingüísticos de comportamiento de los usuarios de Twitter que sufren depresión en comparación con la población general de tuiteros.
Investigadores del Grupo de Informática Biomédica Integrada del GRIB han identificado las características y patrones de comportamiento en los tuits que pueden indicar signos de depresión. El estudio del grupo de Investigación en Informática Biomédica Integrada, publicado en el Journal of Medical Internet Research, es el primero de este tipo en el que se analizan los tuits en lengua española.
El estudio coordinado por Ferran Sanz, ha analizado los patrones lingüísticos y las características de comportamiento de los usuarios de Twitter que sufren depresión en comparación con la población general de tuiteros. "Hemos observado que los usuarios con depresión son más activos en Twitter por la noche y los fines de semana respecto a otros usuarios", detalla Angela Leis, investigadora predoctoral y primera autora del trabajo. Esto podría explicarse como resultado del insomnio, uno de los síntomas más frecuentes de la depresión. Además, añade que "suelen interaccionar menos con otros usuarios, retuiteando menos frecuentemente y con textos más cortos". Asimismo, "su vocabulario se caracteriza por una mayor atención en sí mismos, con un estilo de escritura menos variado en el que suelen aparecer palabras de ideación suicida, con abundancia de palabras de carácter negativo, y son frecuentes emociones como tristeza, enojo y repulsión".
Leer noticia aquí

6/6/2019
A webserver to study the protein networks perturbed in diseases
Researchers from the UPF, IMIM and UVIC have developed the website GUILDify to study the molecular environment of the proteins involved in diseases. The website is a promising tool to study the molecular mechanisms underlying a disease, to understand the relationships between two diseases and to propose potential drugs for their treatment.
Quim Aguirre, PhD student at the Structural Bioinformatics Group (SBI) of GRIB, behind this project, tells us all about it in an article on the El·lipse website.

17/05/2019
A conference to promote women's research in computational biology. Abstract submission deadline: July 1st, 2019!!
BSC, UPC and the GRIB (IMIM-UPF) organize the Advances in Computational Biology conference to be held November 28-29 in La Pedrera (Barcelona).
The first Advances in Computational Biology conference will bring together researchers working on systems biology, omics technologies, artificial intelligence and high-performance computing (HPC) with applications to biology from both the public and the private sectors. The conference will be held November 28-29 in La Pedrera (Barcelona). Mar Albà, head of the Evolutionary genomics group of GRIB is member of the organizing committee and Laura I. Furlong, head of the Integrative Bioinformatics group of GRIB is member of the scientific committee of the event.
Maria Jose Rementeria, Social Link Analytics group leader at the Barcelona Supercomputing Center (BSC) and one of the organizers, states: "One of the main purposes of the conference is to visualize and promote the research done by women scientists and for this reason all presenters will be women, although the conference is open to everyone. We want to create a space to foster collaborations between scientists, providing a unique opportunity to share ideas and build research networks".
The programme will include poster and oral presentations, as well as keynotes from leading scientists in the computational biology and HPC fields. The confirmed keynote speakers are Christine Orengo, group leader of Orengo Group at University College London, Natasa Przulj, group leader of the Life Sciences - Integrative Computational Network Biology at the BSC and Marie-Christine Sawley, director of the Exascale Lab at Intel.

30/05/2019
New approaches of chemical safety assessment discussed between regulatory experts and the researchers of EU-ToxRisk
An international workshop, held in Espoo (Fin), close to the headquarters of the European Chemicals Agency (ECHA), paved the way for a new risk assessment strategy, relying less on animal data, and using multi-dimensional information from many sources in the so-called read-across procedure.
The event took place on 21-22 May 2019 and was organised by EU-ToxRisk, in close collaboration with regulatory representatives from several European agencies (ECHA, EFSA, SCCS), US agencies (NTP, EPA), and other organisations (OECD, Health Canada, NIHS Japan).
EU-ToxRisk - An Integrated European 'Flagship' Program Driving Mechanism-based Toxicity Testing and Risk Assessment for the 21st Century - is a research project receiving funding from the European Union's Horizon 2020 research and innovation programme. Its main goal is to increase confidence in the implementation of non-animal approaches into the regulatory decision-making process for hazard and risk assessment. For this purpose, the project works on the development, evaluation and implementation of animal-free New Approach Methods (NAM) in modern toxicology. GRIB participates on this project, in particular, the PharmacoInformatics group of GRIB is leading the development of computational tools which will facilitate the use of NAM in read accross studies.
09/05/2019
New version of DisGeNET 6.0 and ELIXIR recognition
The Integrative Biomedical Informatics group of GRIB (IMIM-UPF) has launched a new version of DisGeNET, a public knowledge management platform on the genomics of disease, which is celebrating its tenth anniversary this year. DisGeNET offers information on genes and genomic variants associated with human diseases, which is obtained by integrating more than a dozen public resources and the scientific literature. DisGeNET contains one of the most comprehensive collections of genes and variants associated with human diseases that is currently available.
The new version of DisGeNET (6.0) contains approximately 600,000 associations between more than 17,000 genes and 24,000 human diseases, focusing particularly on genetic alterations associated with disease: this version includes more than 117,000 genomic variants associated with 10,000 diseases. In addition, the phenotypic landscape covered by DisGeNET has been expanded to include the genomic basis of clinical manifestations of diseases, both signs and symptoms, as well as laboratory tests results.
Finally, we are pleased to announce that the RDF API, which allows DisGeNET to be exploited in an integrated way with other resources, has been selected as one of the 10 interoperability resources recommended by ELIXIR, the European organisation that brings together bioinformatics resources in life sciences. This recognition is awarded to resources that follow the FAIR (Findable, Accessible, Interoperable and Reusable) principles and which let computers automatically connect resources for complex queries.
29/04/2019
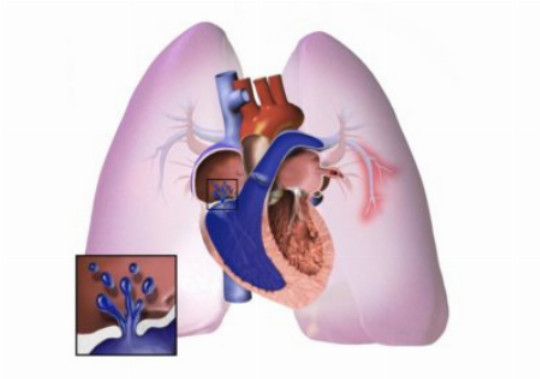
A region in the genome involved in the development of heritable pulmonary arterial hypertension described
A new study has identified a region in the genome that could contribute to increased susceptibility to suffering pulmonary arterial hypertension among carriers of a mutation in the gene BMPR2, which is responsible for the heritable form of the diseas
Researchers from the IDIBAPS, GRIB (IMIM-UPF) and the Israel Institute of Technology, Technion, have identified a region in the genome that could contribute to increased susceptibility to suffering pulmonary arterial hypertension among carriers of a mutation in the gene BMPR2, which is responsible for the heritable form of the disease. The study was published in Journal of Medical Genetics.
Heritable pulmonary arterial hypertension (HPAH) is a rare disease characterized by an abnormal increase in the average blood pressure in the lung. This disorder is mainly caused by mutations in the gene BMPR2. However, identifying mutations in certain genes is often not enough to understand the genetic basis of rare heritable diseases. In fact, some carriers of the disease have been seen not even to develop the disease, a phenomenon known as incomplete penetrance. In recent years this phenomenon has appeared to be far more significant and widespread than had been imagined in previous decades.
"Our task has been to study which genetic modifiers can explain this difference between healthy and diseased carriers", explains Robert Castelo, head of the Functional Genomics group of GRIB. The researchers carried out the study in a family affected by HPAH in which about 40% of the carriers of the mutation have developed the disease. The family studied includes 65 individuals from five different generations, of whom 22 are carriers of the mutation and eight have been diagnosed with HPAH. "By analysing their genetic profiles, we have managed to identify a region in the genome that could potentially contribute to showing susceptibility to the disease among carriers of the mutation", says Pau Puigdevall, first author of the article and researcher of GRIB.

29/04/2019
Congratulations to Alba Gutiérrez Sacristán for receiving one of the UPF Doctoral School PhD Extraordinary Awards and the International PhD Mention.
During the 2017-2018 academic year, a total of 86 PhD theses were read and 9 of them have been awarded
Congratulations to Alba Gutiérrez Sacristán, doctoral student of the Integrative Biomedical Informatics group of GRIB for her thesis "A bioinformatics approach to the study of comorbidity. Insight into mental disorders", directed by Dr. Laura Inés Furlong. She developed a new bioinformatics approach for the exploitation of information contained in clinical health records as well as the identification of comorbidity patterns. Furthermore, by using automatic text mining tools, this thesis also presented PsyGeNET, a database of psychiatric disorders and their genes developed in collaboration with experts in the field of psychiatry and neurosciences. Psychiatric patients frequently present other comorbid diseases, which affect both treatment options and clinical evolution. In this regard, a tool such as PsyGeNET supports the study of the molecular and cellular mechanisms that underpin psychiatric disease comorbidities, providing new insights into new disease biomarkers and drug targets. This thesis has received one of the UPF Doctoral School PhD Extraordinary Awards and also the International PhD Mention.
The PhD Programme in Biomedicine run by the DCEXS currently enrols 456 doctoral students. During the 2017-2018 academic year, a total of 86 PhD theses were read and their corresponding examination committees put 21 of them forward to compete for the PhD Special Awards. The awards ceremony will take place during the 2020 UPF Graduation Ceremony for master's and doctoral degree students.

10/04/2019
Jordi Mestres takes part in the European project EOSC-Life which aims to develop an open collaborative space for digital biology
The project EOSC-Life aims to create an open collaborative digital space for life science in the European Open Science Cloud (EOSC). EOSC-Life brings together the 13 european research infrastructures in the Health and Food domain of the ESFRI Roadmap and is funded by H2020 for the period 2019-2023. The project involves research groups from 46 European academic institutions. These include the GRIB Systems Pharmacology research group (IMIM-UPF) led by Jordi Mestres, which is helping develop collaborative tools for integrating and analysing all kinds of data in the field of life sciences.
According to Dr. Mestres: "Taking part in this macroproject is a unique opportunity to contribute to the development of open, interoperable and reusable computational tools across all strategic research infrastructures in Europe. The potential impact of this initiative is enormous and goes beyond the European level. There is a growing willingness to share data, to properly annotate and integrate it, and to develop tools for analysing it, to facilitate open access and use by the entire scientific community."
Niklas Blomberg, Director of ELIXIR and Coordinator of the project said: "EOSC-Life will bring together the resources, tools, skills and expertise necessary to transform the fragmented research data landscape into an open space for digital biology. Our ultimate goal is to maximise the collective potential of biomedical ESFRI research infrastructures and the researchers using them. By the end of this project EOSC-Life will be established as the new norm for digital biology in Europe - accessible by Europe's 500,000 life scientists."
04/04/2019
Congratulations to the Computational Science group and Acellera for the great results at the D3R Grand Challenge 4
They have ranked first in two of the subchallenges.
The D3R Grand Challenge 4 is an international competition which offers several tasks considered to be of pharmaceutical interest. Usually, these tasks involve predicting how a drug binds into a protein and/or how strong this binding is. Ideally, if one could predict with high accuracy these values, the drug discovery process will be accelerated and the costs of developing new drugs would decrease. Hence, one of the valuable aspects of these challenges is that they can provide a rough measure of how far we are from this ideal situation.
Acellera and the Computational Science group, led by Gianni de Fabritiis, worked together in this project, using some tools which have already been available for a while at PlayMolecule, like KDeep or DeltaDelta, and developing a new one: SkeleDock.
This week, the D3R organizers have announced the results for the different subchallenges of the competition. We are happy to report they have ranked first in two of them (BACE free energy prediction and BACE scoring, both in stage 2).

22/03/2019
“We have created one of the world’s largest database of toxicology data”
Manuel Pastor, head of the PharmacoInformatics group of GRIB, talks about the European eTOX project, in which they have managed to get several pharmaceutical companies to share their toxicology data.
eTOX is a European project funded by the Innovative Medicines Initiative (IMI), which aims to promote the development of new drugs, especially in areas where there are unmet medical or social needs. In this case, the project focuses on drug toxicology. "The value of the eTOX project is to create a culture of collaboration among different pharmaceutical companies, an efficient way to use the information that they generate in a field as important as the toxicology", explains Pastor.
Read the full interview at the last edition of el·lipse magazine



